1. Introduction
In the last two decades, Acinetobacter baumannii has emerged as a major nosocomial pathogen, able to cause significant morbidity and mortality in healthcare settings and almost impossible to eradicate in the hospital environment (Roy et al., 2022). The increasing resistance of A. baumannii to the last-line treatment options is a global problem. Carbapenems used to be the mainstay for the treatment of nosocomial infections caused by A. baumannii (Maragakis & Perl, 2008). Unfortunately, the overuse of carbapenems, especially in the treatment of extended spectrum beta-lactamases producing Enterobacteriaceae, has rapidly resulted in the worldwide dissemination of carbapenem-resistant nonfermentors (Towner et al., 2009). Carbapenem-resistant A. baumannii (CRAB) isolates are, as a rule, multidrug-resistant (MDR) strains and represent a severe epidemiological and therapeutical challenge (Perez et al., 2007). That’s why CRAB is one of the critical-priority pathogens on the World Health Organization’s priority list of antibiotic-resistant bacteria for effective drug development (Tacconelli et al., 2018).
The most prevalent mechanism responsible for carbapenem resistance in A. baumannii is the production of carbapenem-hydrolyzing enzymes, Ambler class D β–lactamases or oxacillinases (OXAs): the intrinsic OXA-51 and the most common acquired OXAs (OXA-23, OXA-24/40, OXA-58 and OXA-143) (Poirel et al., 2010). Chromosomally located carbapenemases can confer higher resistance with the ISAba1- an enhanced expression of the intrinsic chromosomal blaOXA-51 gene (Turton et al., 2006). Less frequent, but much more powerful carbapenem-hydrolyzing enzymes in A. baumannii (VIM, SIM, IMP, NDM) belong to class B β–lactamases or metallo-β-lactamases (MBLs) (Bonin et al., 2012; Poirel & Nordmann, 2006).
In the attempt to monitor the epidemic evolution of CRAB, several genotypic methods exist. The most commonly used typing techniques include PCR-based and sequencing methods [repetitive sequence-based PCR (rep-PCR), multilocus sequence typing (MLST), sequence-based typing and their allele-specific multiplex PCRs], DNA-based fingerprinting methods [pulsed-field gel electrophoresis (PFGE)] and whole genome sequencing (WGS) (Kamolvit et al., 2015). Molecular typing of the CRAB strains from various European hospitals has shown the emergence of three successful clones originally named European clones I to III, which were renamed as international clones or clonal complexes (ICs/CCs) I to III, after being identified worldwide (Zarrilli et al., 2013).
Although Serbia is a country with a significant trend of rising prevalence of carbapenem-resistant Acinetobacter spp., from 95.1% in 2017 to 98% in 2021 (ECDC, 2023), there is no systematic data or continuous monitoring of the occurrence of CRAB isolates. The present review aims to go through the molecular epidemiology of CRAB clinical isolates in Serbia, according to current published data, and attempt to identify future challenges and trends.
2. Carbapenem-resistant A. baumannii in Serbia
2.1. Resistance rates
CRAB isolates were first recorded in our hospitals in the early 2000s. The resistance to imipenem rose remarkably from 6.9% in 2002 to 67.4% in 2010. Since resistance to third and fourth-generation cephalosporins as well as ciprofloxacin reached 100% in 2010, strains were characterized as MDR or ‘nonsusceptible to at least one agent in ≥3 antimicrobial categories’, according to proposed criteria (Medić et al., 2011). From the early beginnings, the prevalence of carbapenem-resistant Acinetobacter spp. in Serbian hospitals reached 98% in 2021 (ECDC, 2023).
2.2. Molecular epidemiology
Despite the increasing prevalence of carbapenem resistance, data about the molecular epidemiology of CRAB in Serbia is lacking. However, there were reports of OXA and MBL-positive CRAB strains, recovered from patients migrating from Serbia to Western Europe.
German authors reported the molecular characterization of MDR A. baumannii collected from a patient with a femorocrural dacron bypass infection admitted to the intensive care unit at the Frankfurt University Hospital in 2007, after hospitalization in Serbia. The strain was resistant to β-lactams (including carbapenems), fluoroquinolones, aminoglycosides, tigecycline and aztreonam. It remained susceptible to colistin. Isolate harboured blaNDM-1 and intrinsic blaOXA-64 gene, without the insertion sequence ISAba1 located upstream. PCR-based A. baumannii typing in combination with ApaI PFGE analysis confirmed that it was not related to European clonal lineages 1–3 (Göttig et al., 2010; Pfeifer et al., 2011).
The following year brought another record on MDR A. baumannii isolate recovered from a patient hospitalized in Geneva University Hospitals, Switzerland, from 2009 to 2010, after a transfer from Serbia. A. baumannii was recovered from rectal swabs. It was resistant to all β-lactams (including carbapenems), gentamicin, amikacin, chloramphenicol, tetracycline, and fluoroquinolones and remained susceptible to tobramycin and netilmicin, with minimum inhibitory concentrations (MICs) of colistin, rifampin, and tigecycline being at 0.5, 1, and 1 μg/ml, respectively. PCR and sequencing revealed that A. baumannii coharboured blaOXA-23 and blaNDM-1 genes. MLST was performed, following the Institut Pasteur scheme, and showed that the isolate belonged to the sequence type 1 (ST1) (Poirel et al., 2012).
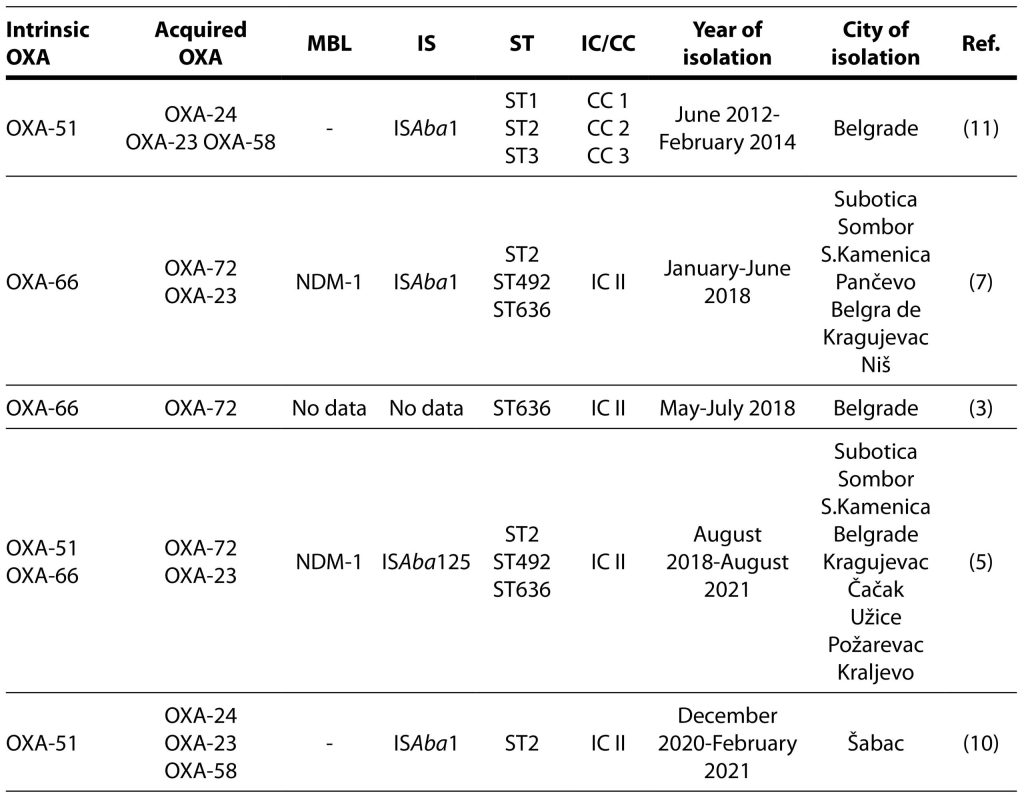
Three years later, the first study from Serbia reported the distribution of carbapenemases among clinical isolates of A. baumannii from a single pediatric hospital. Twenty-eight consecutive, non-duplicate MDR and carbapenem-resistant A. baumannii clinical isolates were collected from June 2012 to February 2014 at the Institute for Mother and Child Health Care “Dr. Vukan Čupić”, a tertiary care paediatric hospital in Belgrade. The majority of isolates (67.68%) belonged to the ST1, European CC II. All isolates harbored intrinsic OXA-51 carbapenemase, while OXA-24, OXA-23, and OXA-58 were detected in 82.14%, 57.14%, and 39.29%, respectively. This study did not detect β-lactamases OXA-143, OXA-235, NDM-1, VIM and IMP. ISAba1 was present upstream of OXA-51 in one isolate, and upstream of OXA-23 in 10 isolates (Novovic et al., 2015).
Molecular epidemiology of 222 CRABs recovered from inpatients with confirmed bacterial infections admitted at nine hospitals throughout Serbia (General hospitals in Subotica, Pančevo and Sombor, Institute for Pulmonary Diseases of Vojvodina, University Hospital Medical Center Bežanijska kosa, University Hospital Center dr Dragiša Mišović, Institute for Cardiovascular Diseases Dedinje, Clinical Center Kragujevac, Clinical Center Niš) during the period January–June 2018 was published as a part of the prospective, observational, multicenter study in 2020. All isolates carried the naturally occurring blaOXA-51 gene, while blaOXA-24 and blaOXA-23 were detected in 44.2%, and 34.5% CRABs, respectively. However, blaOXA-58 and blaOXA-143 genes were not discovered. Overall, ISAba1 was present in 71.8% of CRABs. The blaNDM-1 was the only MBL gene detected in the study. None of the isolates carried blaIMP, blaVIM, blaGIM, blaSPM and blaSIM genes. There were no substantial differences between the hospitals and regions regarding the proportion of isolates carrying different acquired blaOXA genes. However, blaNDM-1 positive isolates were detected only in Belgrade and Niš. Sequencing of the blaOXA genes revealed the presence of the blaOXA-66, blaOXA-72, and blaOXA-23 variants. MLST assigned the analyzed CRABs to 3 STs: ST2, ST492 (a single locus variant of ST2), and ST636 (a triple locus variant of ST2). ST2 and ST492 belonged to ICII, while ST636 was a singleton, not categorized in any IC (Lukovic et al., 2020).
The following year, Serbian authors reported an A. baumannii outbreak among preterm neonates in a neonatal intensive care unit at the Institute of Neonatology in Belgrade. During the outbreak period (May-July 2018), there were 13 cases of A. baumannii bloodstream infection among 82 hospitalized neonates. All A. baumannii strains were carbapenem-resistant and susceptible to colistin. Molecular characterization of the isolates revealed that they harboured OXA-66 and OXA-72 β-lactamases and belonged to ST636 (ICII), while the PFGE pattern indicated clonal spread. Lower gestational age, lower Apgar score, vaginal delivery and mechanical ventilation were risk factors for A. baumannii infection. Four patients died, eight patients were treated successfully with colistin, and one patient with sepsis and meningitis on dual ampicillin-sulbactam and colistin therapy recovered with sequelae. The outbreak was eventually controlled by reinforcement of the infection control measures based on a multi-tiered interventional approach (Gajic et al., 2021).
Kabic et al. (2022) analyzed comparative genomics and molecular epidemiology of 30 colistin-resistant A. baumannii isolated from clinical specimens collected from patients admitted to the hospitals in nine cities throughout Serbia between August 2018 and August 2021. All isolates were CRAB and were resistant or had reduced susceptibility to amoxicillin-clavulanic acid, piperacillin-tazobactam, ceftazidime, cefepime, cefoxitin, meropenem, imipenem, amikacin, gentamicin, tobramycin, levofloxacin, ciprofloxacin, and tetracycline. Three STs were identified based on the Pasteur MLST scheme. Among them, ST2 was the most prevalent (76.66%), followed by ST492 (20%), and ST636 (3.33%). Several β-lactamase-encoding genes were detected. Overall, 76.66% of isolates carried the blaOXA-23 gene, while 23.33% and 10 % of isolates harboured the blaOXA-72 and blaNDM-1 as single-copy genes, respectively. Additionally, ISAba125 was located upstream of the blaNDM-1 gene.
Since the World Health Organization declared the COVID-19 pandemic in March 2020, the disease has spread rapidly, leading to an overload of the health system, and many of the patients infected with SARS-CoV-2 needed to be admitted to the intensive care units (ICUs). The 64 A. baumannii isolates were recovered from COVID-19 patients admitted to ICU at General Hospital “Dr Laza K. Lazarevic” in Šabac, during the period from December 2020 to February 2021. All patients required mechanical ventilation, and the mortality rate was 100%.
A. baumannii isolates were sensitive to colistin, while resistant to meropenem, imipenem, gentamicin, tobramycin, and levofloxacin according to the broth microdilution method and MDR phenotype was confirmed. Typically for A. baumannii, intrinsic genes encoding for oxacillinase OXA-51 were detected in all tested isolates by PCR method. Additionally, all isolates gave a positive PCR signal for the blaOXA-23 gene and ISAba1 upstream of this gene. The gene encoding for OXA-24 oxacillinase was identified in 28.12% of isolates, while the blaOXA-58 gene was found in 6.25% of isolates. ISAba1 was not detected upstream of blaOXA-24, blaOXA-51, and blaOXA-58 genes in analyzed isolates. The blaNDM-1 gene was not present in the genomes of tested isolates. MLST analyses revealed that all isolates obtained from Šabac hospital were identified as ST2 (IC II) according to Pasteur nomenclature (Novović et al., 2023).
Carbapenem-hydrolyzing enzymes, IS, ST and IC connected to CRAB strains isolated in Serbia are shown in Table 1.
Table 1. Carbapenem-hydrolyzing enzymes isolated in carbapenem-resistant Acinetobacter baumannii strains in Serbia.
OXA-oxacillinase; MBL- metallo-β-lactamase; IS-insertion sequence; ST-sequence type; IC-international clone; CC-clonal complex
3. Conclusion
In summary, this study highlights the molecular epidemiology of the CRAB clinical isolates. Since their first occurrence around 2000, CRAB strains have become endemic in Serbia. Their emergence was associated with carbapenem consumption, while the percentage for carbapenem resistance reached 98% in 2021. OXA carbapenemases constitute the most prevalent mechanism of resistance. CRABs harboring blaOXA-51, blaOXA-23 and blaOXA-24 genes are dominant. The prevalence of OXA-23 and OXA-24 producing CRAB may have occurred due to their higher hydrolytic activity, compared with OXA-58 producers. The higher MICs to carbapenems possibly provided them a comparative advantage to survive and prevail in the nosocomial setting.
This review states that Serbia might be an endemic region for CRAB isolates carrying blaNDM-1 genes. NDM-1-producing CRABs were recovered in our hospitals and also from patients migrating from Serbia to Western Europe.
There are several major epidemic lineages, including ST1 (CC II), ST2 (IC II), ST492 (IC II), and ST636, with various acquired carbapenemase genes, usually associated with a plasmid and transposable mobile genetic elements in these strains, which could explain the horizontal transfer of blaOXA and blaMBL across different lineages.
The extremely high antimicrobial resistance rates and the emergence of specific CRAB clones point out the critical need for the implementation of continuous surveillance of infections caused by A. baumannii and the development of accurate prevention strategies in Serbia.
Funding: This research was supported by the Science Fund of the Republic of Serbia, #GRANT No 7042, Tracking antimicrobial resistance in diverse ecological niches – one health perspective – TRACE] and The Ministry of Science, Technological Development and Innovation of the Republic of Serbia, #GRANT No 451-03-66/2024-03/200110, subgrant Multidrug-Resistant Bacteria in the Hopsital Environment in Serbia, Prevalence, Circulating clones, Genetic Basis of Antibiotic Resistance and Virulence Factors.
Informed Consent Statement: Not applicable.
Data Availability Statement: Not applicable.
Conflicts of Interest: The authors declare no conflicts of interest.
References
Bonnin, R. A., Poirel, L., Naas, T., Pirs, M., Seme, K., Schrenzel, J., & Nordmann, P. (2012). Dissemination of New Delhi metallo-β-lactamase-1-producing Acinetobacter baumannii in Europe. Clinical microbiology and infection: the official publication of the European Society of Clinical Microbiology and Infectious Diseases, 18(9), E362–E365.
European Centre for Disease Prevention and Control (ECDC) and World Health Organization (n.d.). 2023. Antimicrobial resistance surveillance in Europe 2023 – 2021 data. https://www.ecdc.europa.eu/en/publications-data/antimicrobial-resistance-surveillance-europe-2023-2021-data. Accessed on: 14.04.2024
Gajic, I., Jovicevic, M., Milic, M., Kekic, D., Opavski, N., Zrnic, Z., Dacic, S., Pavlovic, L., & Mijac, V. (2021). Clinical and molecular characteristics of OXA-72-producing Acinetobacter baumannii ST636 outbreak at a neonatal intensive care unit in Serbia. The Journal of hospital infection, 112, 54–60.
Göttig, S., Pfeifer, Y., Wichelhaus, T. A., Zacharowski, K., Bingold, T., Averhoff, B., Brandt, C., & Kempf, V. A. (2010). Global spread of New Delhi metallo-β-lactamase 1. The Lancet. Infectious diseases, 10(12), 828–829.
Kabic, J., Novovic, K., Kekic, D., Trudic, A., Opavski, N., Dimkic, I., Jovcic, B., & Gajic, I. (2022). Comparative genomics and molecular epidemiology of colistin-resistant Acinetobacter baumannii. Computational and structural biotechnology journal, 21, 574–585.
Kamolvit, W., Sidjabat, H. E., & Paterson, D. L. (2015). Molecular Epidemiology and Mechanisms of Carbapenem Resistance of Acinetobacter spp. in Asia and Oceania. Microbial drug resistance (Larchmont, N.Y.), 21(4), 424–434.
Lukovic, B., Gajic, I., Dimkic, I., Kekic, D., Zornic, S., Pozder, T., Radisavljevic, S., Opavski, N., Kojic, M., & Ranin, L. (2020). The first nationwide multicenter study of Acinetobacter baumannii recovered in Serbia: emergence of OXA-72, OXA-23 and NDM-1-producing isolates. Antimicrobial resistance and infection control, 9(1), 101.
Magiorakos, A. P., Srinivasan, A., Carey, R. B., Carmeli, Y., Falagas, M. E., Giske, C. G., Harbarth, S., Hindler, J. F., Kahlmeter, G., Olsson-Liljequist, B., Paterson, D. L., Rice, L. B., Stelling, J., Struelens, M. J., Vatopoulos, A., Weber, J. T., & Monnet, D. L. (2012). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clinical microbiology and infection : the official publication of the European Society of Clinical Microbiology and Infectious Diseases, 18(3), 268–281.
Medić, D., Ukropina, M. M., Gusman, V., Jelesić, Z., & Milosavljević, B. (2011). Medicinski pregled, 64(11-12), 583–587.
Novović, K., Kuzmanović Nedeljković, S., Poledica, M., Nikolić, G., Grujić, B., Jovčić, B., Kojić, M., & Filipić, B. (2023). Virulence potential of multidrug-resistant Acinetobacter baumannii isolates from COVID-19 patients on mechanical ventilation: The first report from Serbia. Frontiers in microbiology, 14, 1094184.
Novovic, K., Mihajlovic, S., Vasiljevic, Z., Filipic, B., Begovic, J., & Jovcic, B. (2015). Carbapenem-resistant Acinetobacter baumannii from Serbia: revision of CarO classification. PloS one, 10(3), e0122793.
Perez, F., Hujer, A. M., Hujer, K. M., Decker, B. K., Rather, P. N., & Bonomo, R. A. (2007). Global challenge of multidrug-resistant Acinetobacter baumannii. Antimicrobial agents and chemotherapy, 51(10), 3471–3484.
Pfeifer, Y., Wilharm, G., Zander, E., Wichelhaus, T. A., Göttig, S., Hunfeld, K. P., Seifert, H., Witte, W., & Higgins, P. G. (2011). Molecular characterization of blaNDM-1 in an Acinetobacter baumannii strain isolated in Germany in 2007. The Journal of antimicrobial chemotherapy, 66(9), 1998–2001.
Poirel, L., Bonnin, R. A., Boulanger, A., Schrenzel, J., Kaase, M., & Nordmann, P. (2012). Tn125-related acquisition of blaNDM-like genes in Acinetobacter baumannii. Antimicrobial agents and chemotherapy, 56(2), 1087–1089.
Poirel, L., Naas, T., & Nordmann, P. (2010). Diversity, epidemiology, and genetics of class D beta-lactamases. Antimicrobial agents and chemotherapy, 54(1), 24–38.
Poirel, L., & Nordmann, P. (2006). Carbapenem resistance in Acinetobacter baumannii: mechanisms and epidemiology. Clinical microbiology and infection : the official publication of the European Society of Clinical Microbiology and Infectious Diseases, 12(9), 826–836.
Roy, S., Chowdhury, G., Mukhopadhyay, A. K., Dutta, S., & Basu, S. (2022). Convergence of Biofilm Formation and Antibiotic Resistance in Acinetobacter baumannii Infection. Frontiers in medicine, 9, 793615.
Tacconelli, E., Carrara, E., Savoldi, A., Harbarth, S., Mendelson, M., Monnet, D. L., Pulcini, C., Kahlmeter, G., Kluytmans, J., Carmeli, Y., Ouellette, M., Outterson, K., Patel, J., Cavaleri, M., Cox, E. M., Houchens, C. R., Grayson, M. L., Hansen, P., Singh, N., Theuretzbacher, U., … WHO Pathogens Priority List Working Group (2018). Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. The Lancet. Infectious diseases, 18(3), 318–327.
Towner, K. J., Levi, K., Vlassiadi, M., & ARPAC Steering Group (2008). Genetic diversity of carbapenem-resistant isolates of Acinetobacter baumannii in Europe. Clinical microbiology and infection : the official publication of the European Society of Clinical Microbiology and Infectious Diseases, 14(2), 161–167.
Turton, J. F., Ward, M. E., Woodford, N., Kaufmann, M. E., Pike, R., Livermore, D. M., & Pitt, T. L. (2006). The role of ISAba1 in expression of OXA carbapenemase genes in Acinetobacter baumannii. FEMS microbiology letters, 258(1), 72–77.
Zarrilli, R., Pournaras, S., Giannouli, M., & Tsakris, A. (2013). Global evolution of multidrug-resistant Acinetobacter baumannii clonal lineages. International journal of antimicrobial agents, 41(1), 11–19.